|
PORECAMP USA |
|
Additional Information
Logistics
- Arrival – On arrival on Monday please contact Charles Johnson
- Storing your DNA on arrival – If you are bringing your own samples we will have an ice bucket out during the reception and will transfer these to a lab fridge. Please ensure your sample is labelled clearly. We can accept no responsibility for loss or damage to your samples. You are welcome to ship your samples early. Please email Emma Schultz to get shipping information.
- Information about the locations and what to bring [PPTX] and [PDF]
Frequently Asked Questions
What to bring
- Yourself
- A modern laptop (this will not be used for MinIon sequencing, but rather to connect to resources to analyze MinIon data). Just in case, please also bring an Ethernet cable and adaptor if required.
- Your favorite lab coat if you have one (we will be able to provide one if needed)
- Pens, markers and something suitable for use as a lab-book
What will we be sequencing?
- We will be using the latest R9 flowcells and preparing barcoded libraries to run across approximately 3 flowcells each day.
How much DNA do we need to bring?
- 1-2ug
Can we bring multiple samples or just two?
- You are welcome to bring more then one sample just in case one fails to meet our QC standards, but you will only be running one as 1 or 2 /12 of a flow cell.
Will we each be using our own computer for the actual sequencing runs?
- We will be suppling computers to do the sequencing runs and you will be logging into a virtual machine to do your bioinformatics. So the laptop is really just for terminal use, taking notes, looking up protocols and such
Can I bring my own material?
- We are happy for participants to bring one sample of their own material for sequencing provided it is genomic. However, please do not rely on the course to obtain results for your experiment. Flowcells are limited and it is unlikely that you will generate sufficient data for your experiment.
- If you do wish to bring your own material, please ensure you prepare no more than two slides outlining what the sample is and why it is interesting. We will need a minimum of around 1ug of material. One your samples arrive we will QC them. We will assess the QC data as part of the course.
- Again, we reiterate – please do not rely on this course to generate data for your experiment. We view the QC exercise as important to demonstrate the range of DNA quality and its impact on library prep and sequencing rather than to ensure that we obtain a good library and data from a given sample.
- If you are on campus then bring it over this week (May 29th to June 2nd) , for everyone else we’ll have an ice bucket at check in. Please make sure they are well labeled. We will QC everyone’s samples Monday afternoon while you are in class.
- We will have DNA that you can use to learn library prep if you don’t bring your own.
Is breakfast/lunch/dinner included?
- Breakfast is not provided. For those staying in the dorms there is a Starbucks
- Lunch is provided everyday
- Dinner is provided only for Monday and Friday. The other weekdays dinner is not provided. This is primarily to give participants the widest possible choice of options.
Is WiFi available?
- Everyone who is not from TAMU will be getting their own guest account that is good for a month, that works in any university building and even outside for most of the areas of campus. Our building has ~250-300Mbps
Papers
- Sequencing and assembly of a human genome with ultra-long reads – 2017 – [BioArxiv] [GenomeWeb)
- Reconstructing The Gigabase Plant Genome Of Solanum pennellii – 2017 – [BioArxiv]
- Reading canonical and modified nucleotides in 16S ribosomal RNA using nanopore direct RNA sequencing [BioArxiv]
- Real-Time DNA Sequencing in the Antarctic Dry Valleys – 2017 – [JBT]
- Delivery of Nanopore sequencing to the genomics community – 2016 – [Genome Biology]
- Benchmarking for quantitative and qualitative assessment of cDNA populations – 2016 – [Scientific Reports]
- More publications here
Location and Maps
Maps
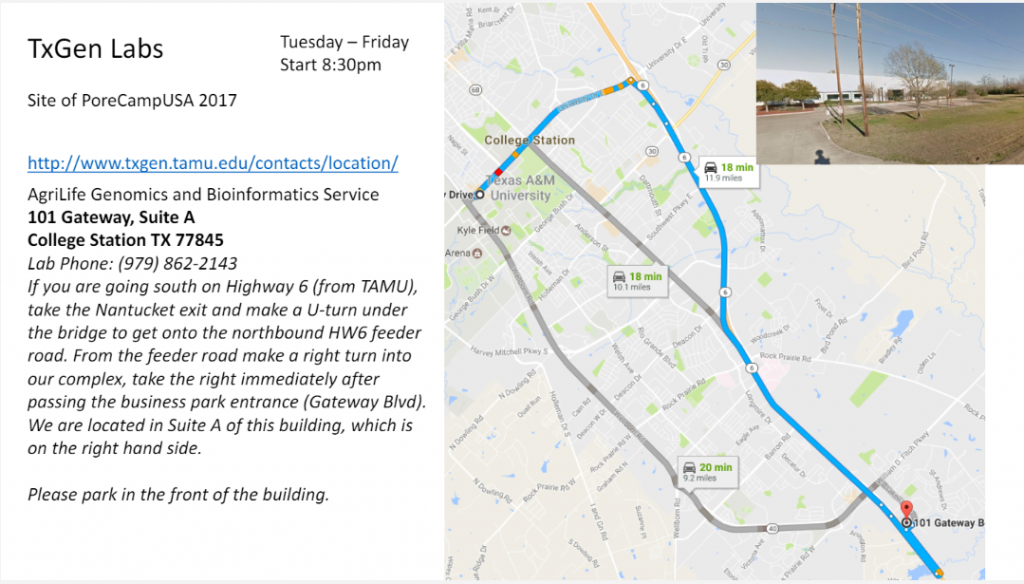
TxGen Lab |
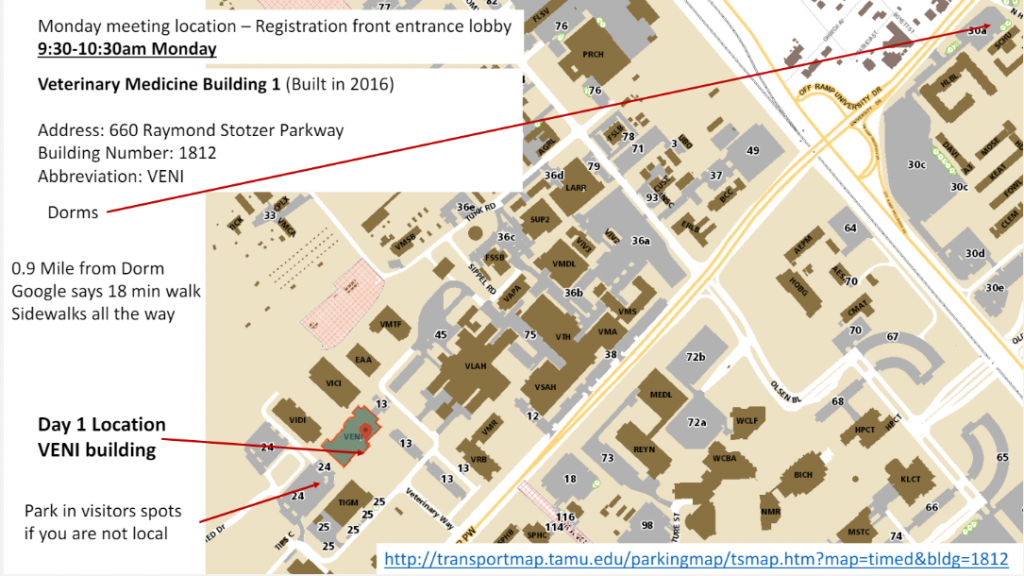
Veterinary Medicine Building |
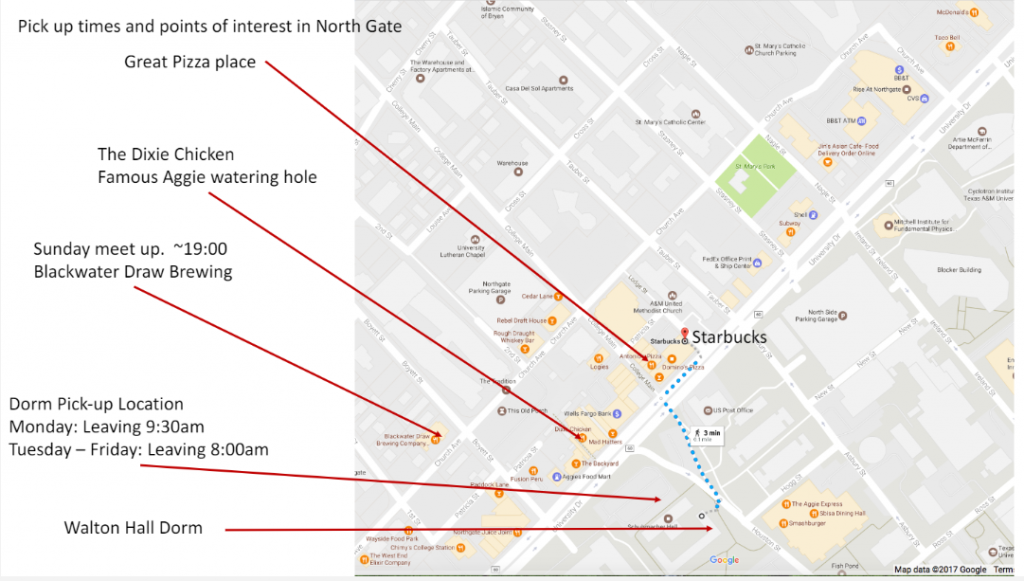
North Gate |
Map to the Lab
- TxGen : 101 Gateway Blvd., Suite A, College Station, Texas
[mappress mapid=”2″]
Sponsors
   |
   |
  |
Acknowledgements
- This work used the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by National Science Foundation grant number ACI-1548562 [1].
- We would like to thank the Texas Advanced Computing Power, https://www.tacc.utexas.edu/, CyVerse, http://www.cyverse.org/, and Jetstream (https://jetstream-cloud.org/) for allocations, donated capacity, and technical assistance.
[1] John Towns, Timothy Cockerill, Maytal Dahan, Ian Foster, Kelly Gaither, Andrew Grimshaw, Victor Hazlewood, Scott Lathrop, Dave Lifka, Gregory D. Peterson, Ralph Roskies, J. Ray Scott, Nancy Wilkins-Diehr, “XSEDE: Accelerating Scientific Discovery“, Computing in Science & Engineering, vol.16, no. 5, pp. 62-74, Sept.-Oct. 2014, doi:10.1109/MCSE.2014.80

